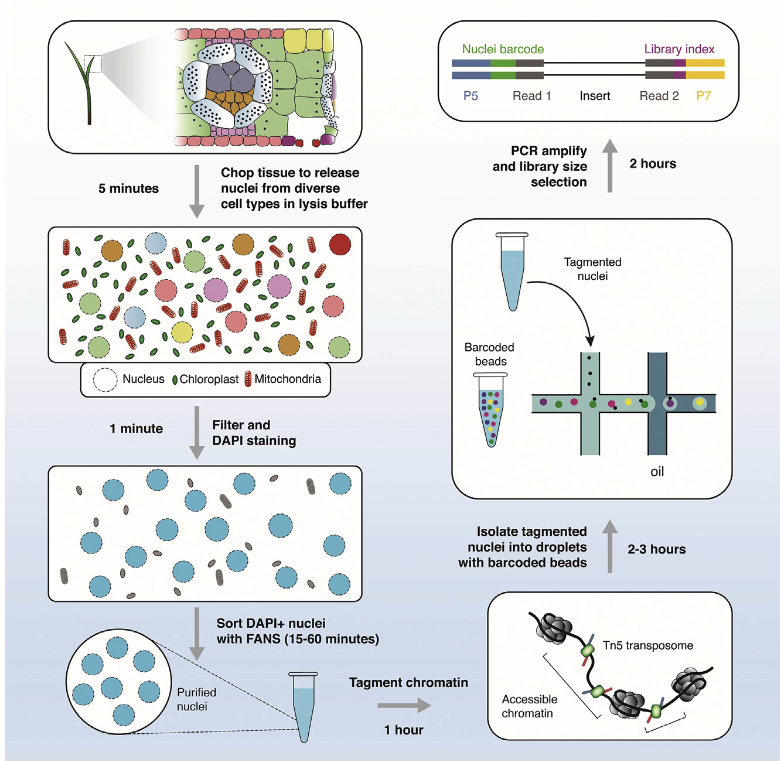
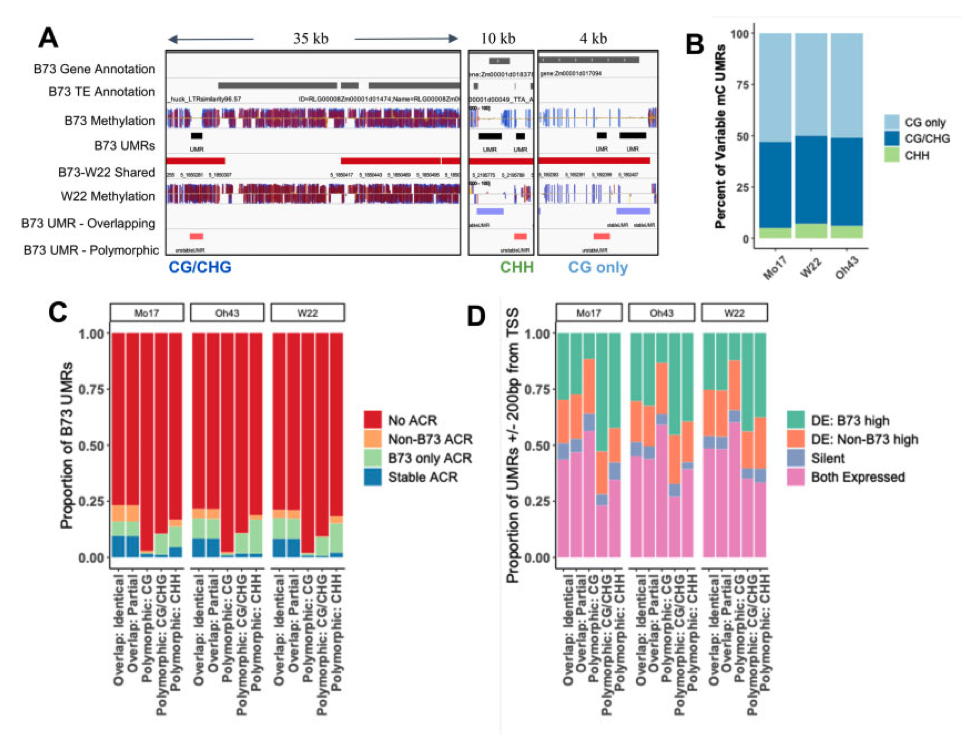
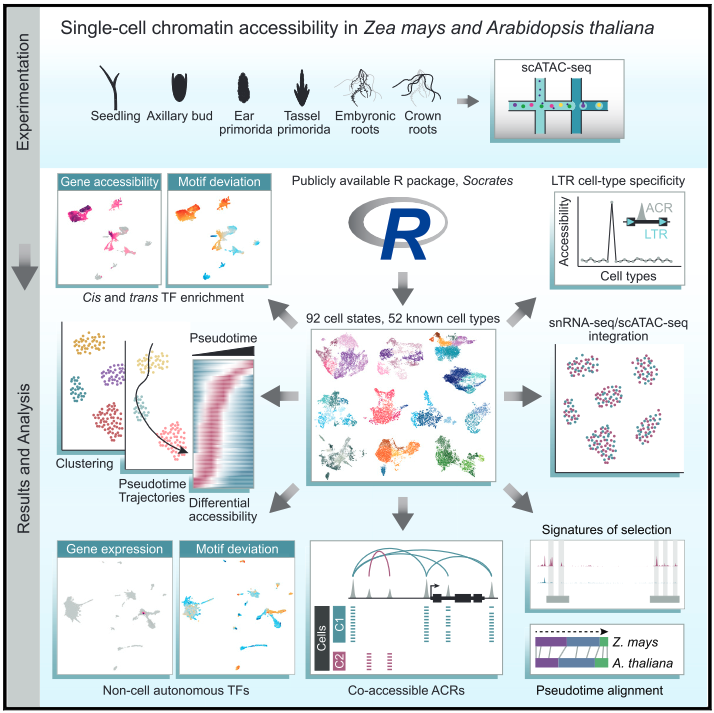
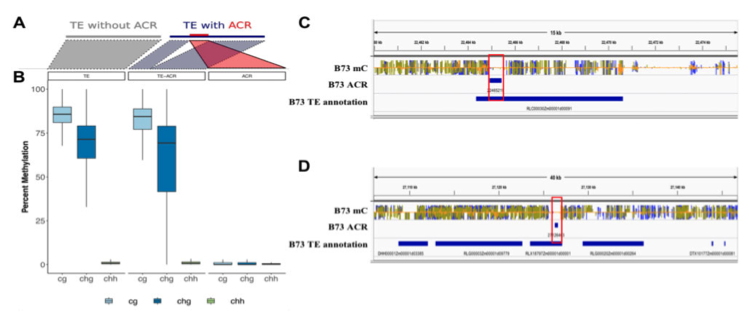
Publications and preprints tracked by Google Scholar can be found here: Google Scholar link
* denotes co-first authors
# denotes (co)corresponding author(s)
2025
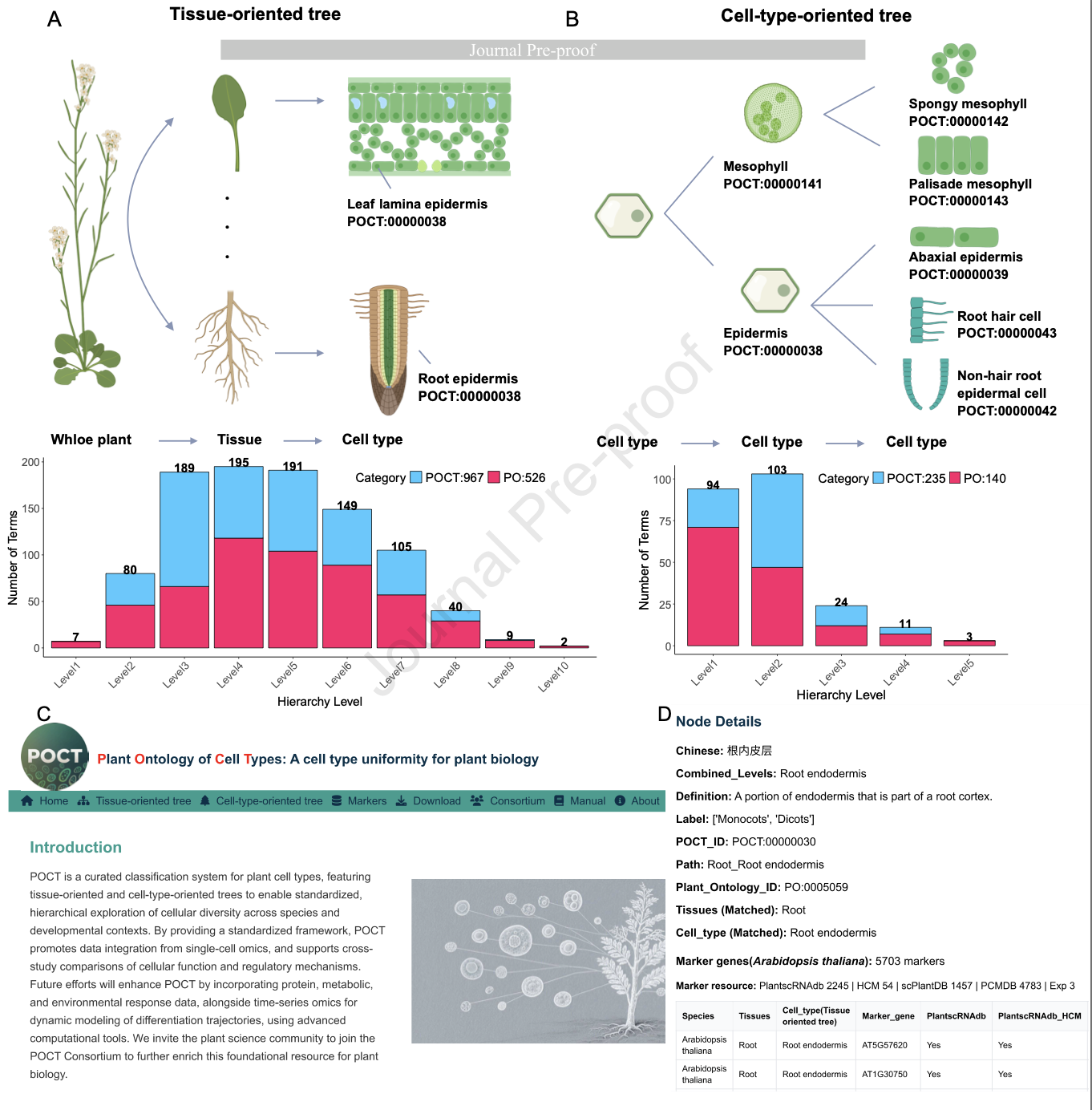
39. The plant ontology of cell types
Hongyu Chen, Pu Liang, Xiaolin Lu, Shuai Jiang, Jingjing Jin, Jiaqi Cai, Yaqian Lu, Dihuai Zheng, Jie Yao, Qinjie Chu, Huixia Shou, Chengxin Fu, Tingting Lu, Yi Jing, Yinqi Bai, Ning Yang, Silin Zhong, Jun Xiao, Fang Yang, Xing Guo, Jixian Zhai, Alexandre P. Marand, Zhixi Tian, Fan Chen, Jian Xu, Yuling Jiao, Dijun Chen, Peijian Cao, Xiaofeng Cui, Dave Jackson, Tatsuya Nobori, Xiaofeng Gu, Jiawei Wang, James Whelan, Jianbing Yan, Robert J. Schmitz, Zhang Zhang, Longjiang Fan, POCT Consortium (2025)
Molecular Plant
38. Widespread turnover of a conserved cis-regulatory code across 589 grass species
Charles O. Hale, Sheng-Kai O. Hsu, Jingjing Zhai, Aimee J. Schulz, Taylor AuBuchon-Elder, Germano M. F. Costa-Neto, Allen Gelfond, Mohamed Z. El-Walid, Matthew Hufford, Elizabeth A. Kellogg, Thuy La, Alexandre P. Marand, Arun S. Seetharam, Armin Scheben, Michelle C. Stitzer, Travis Wrightsman, Cinta Romay, Edward S. Buckler (2025)
Molecular Biology and Evolution
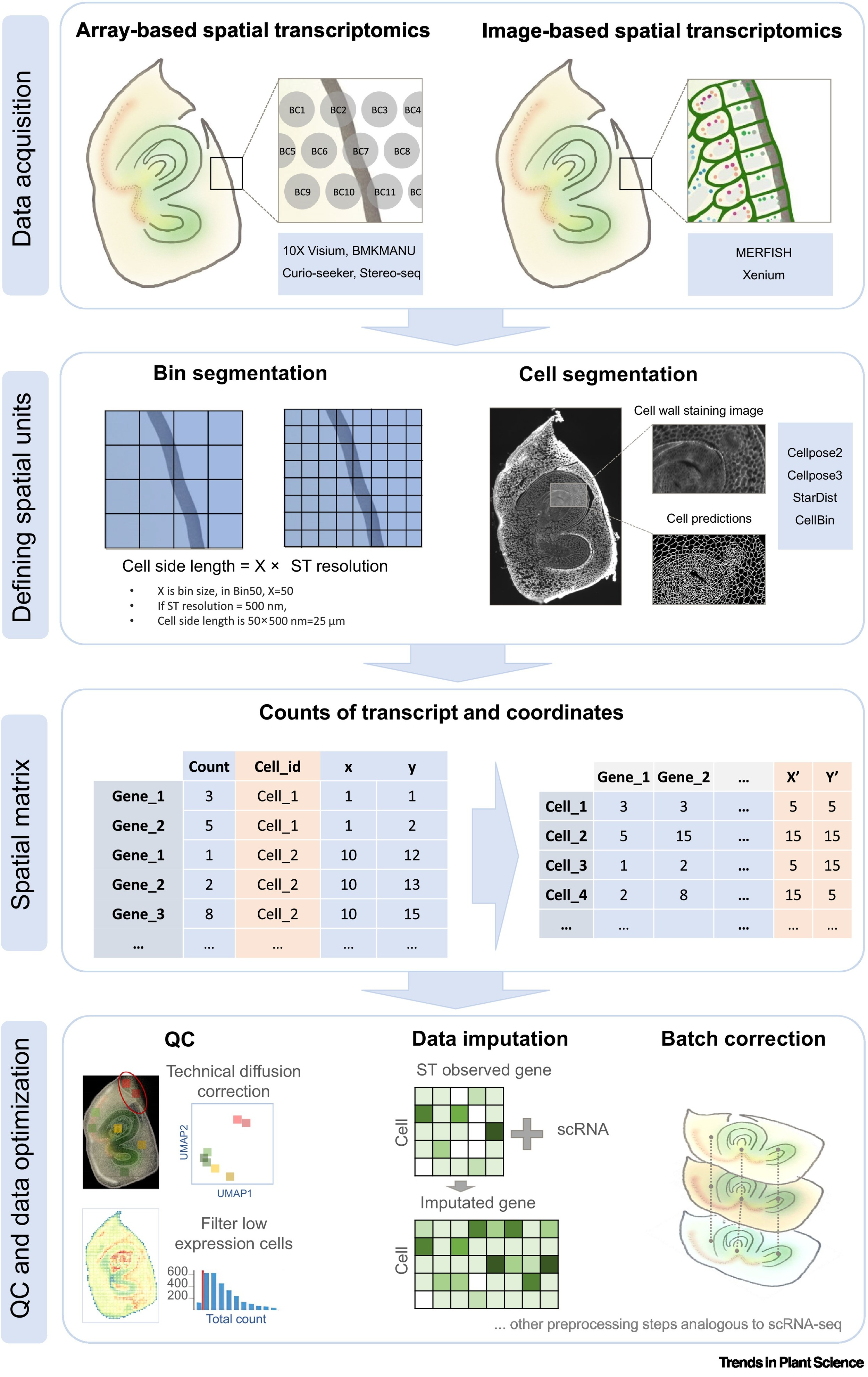
37. Advances in plant spatial multi-omics data analysis
Jie Yao, Alexandre P. Marand, Yinqi Bai, Robert J. Schmitz, Longjiang Fan (2025)
Trends in Plant Science
36. Hybrids reveal accessible chromatin trans genetic associations
Mark A. A. Minow, Alexandre P. Marand, Xuan Zhang, Cullan A. Meyer, Jordan Pelkman, Robert J. Schmitz (2025)
bioRxiv
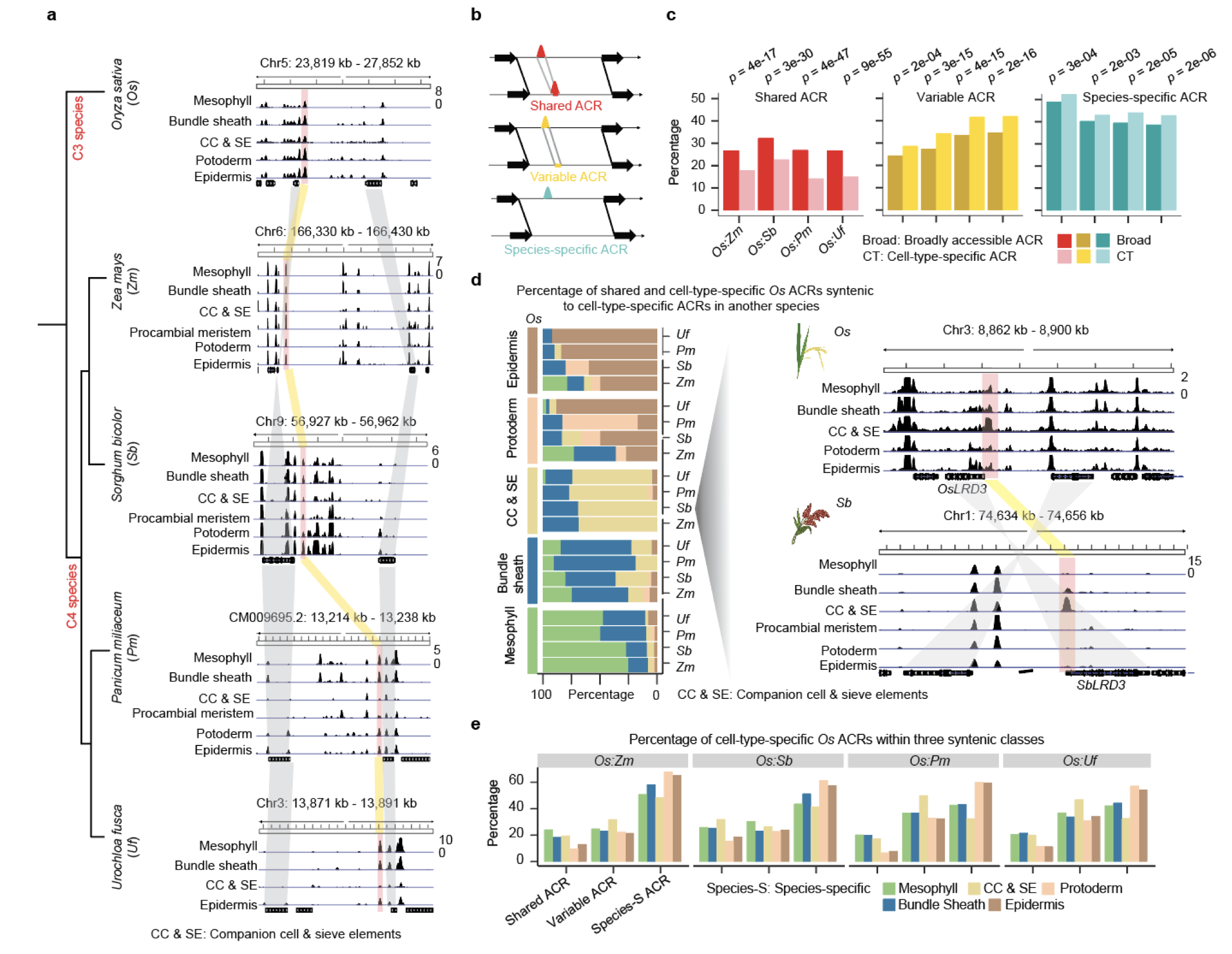
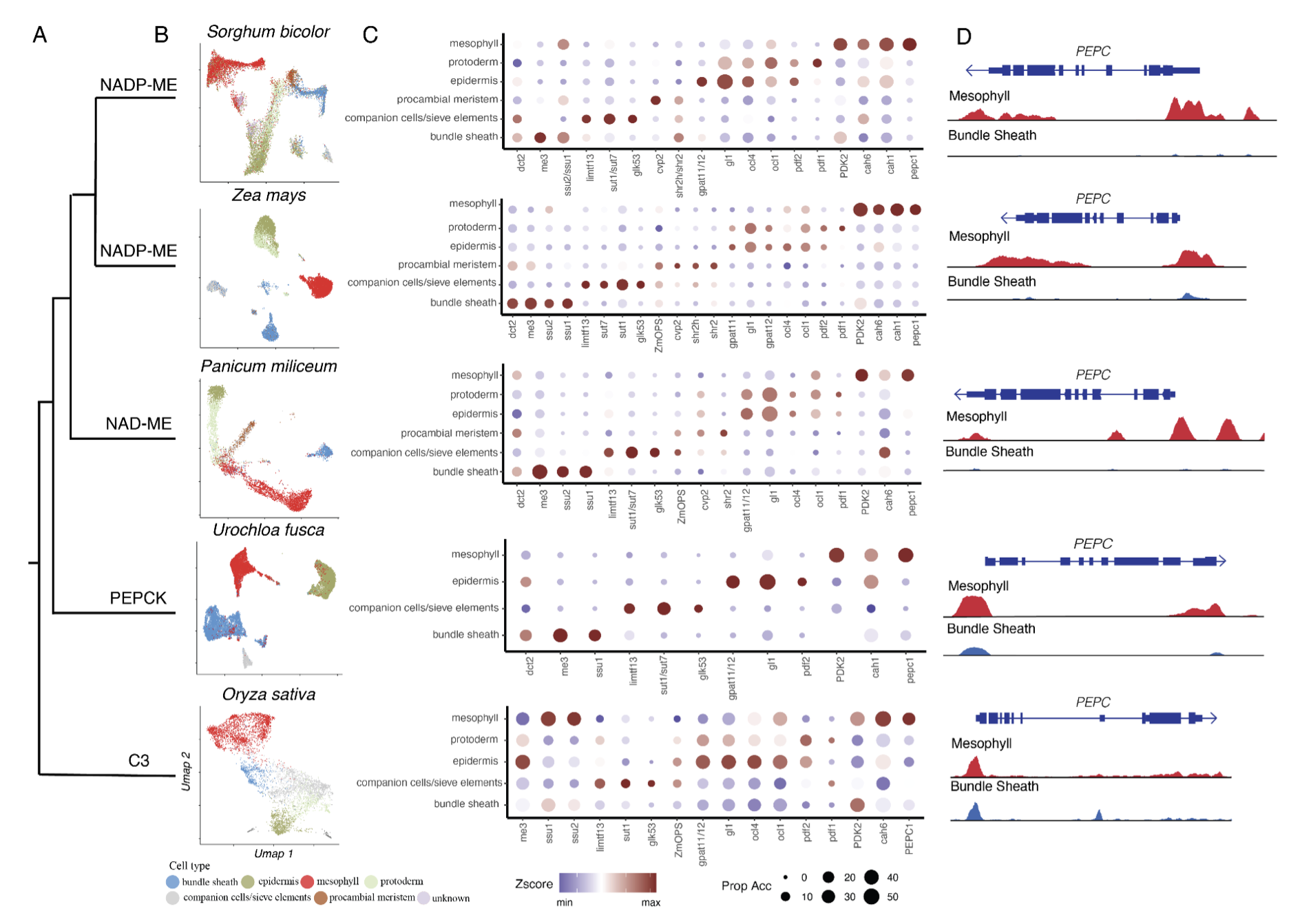
35. A single-cell rice atlas integrates multi-species data to reveal cis-regulatory evolution
Haidong Yan*, John P. Mendieta*, Xuan Zhang, Ziliang Luo, Alexandre P. Marand, Yan Liang, Mark A. A. Minow, Yun Zhong, Hosung Jang, Xiang Li, Thomas Roulé, Doris Wagner, Xiaoyu Tu, Yonghong Wang, Daiquan Jiang, Silin Zhong, Linkai Huang, Susan R. Wessler, Robert J. Schmitz (2025)
Nature Plants
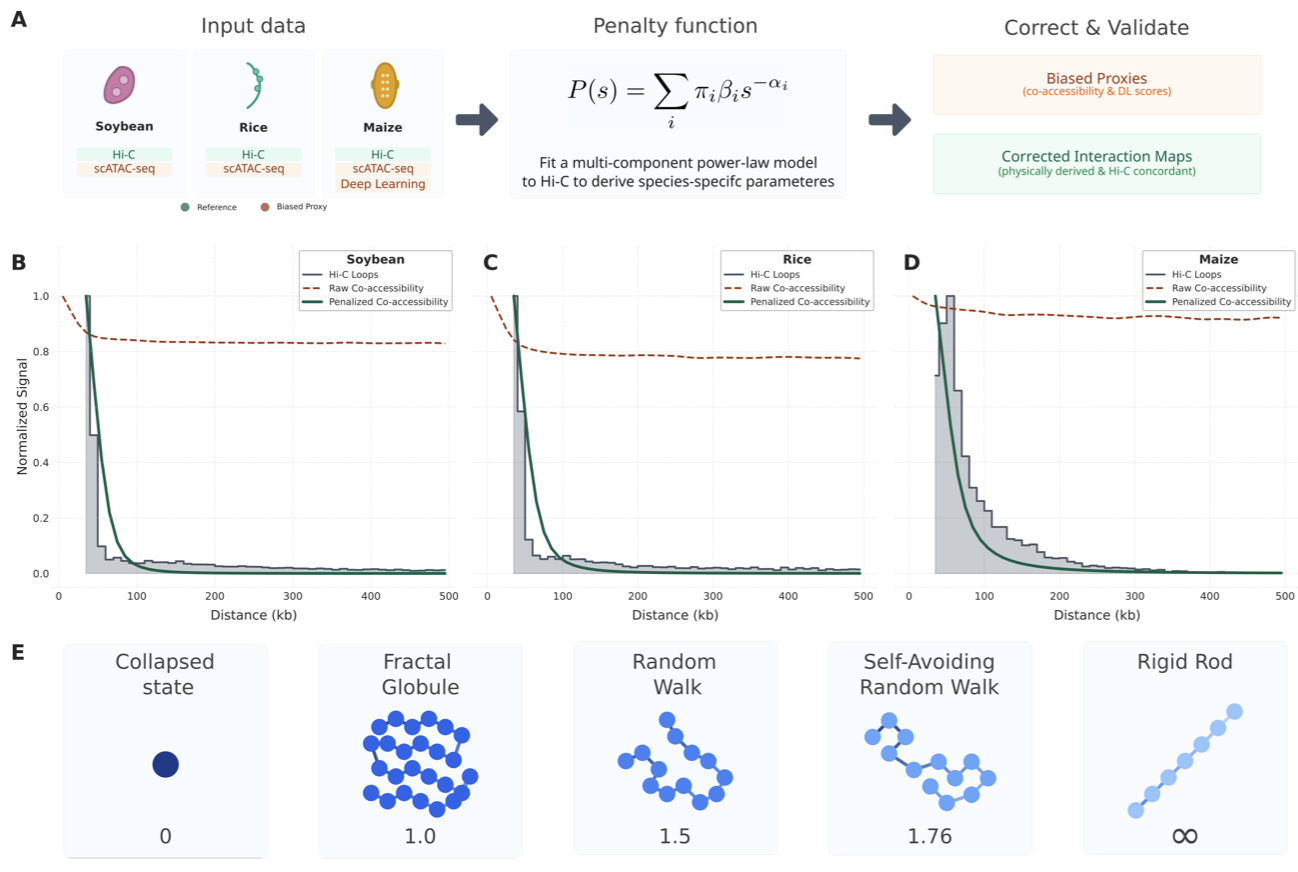
34. Polymer-derived distance penalties improve chromatin interaction predictions from single-cell data across crop genomes
Luca Schlegel, Fabio Gomez-Cano, Alexandre P. Marand#, Frank Johannes# (2025)
bioRxiv
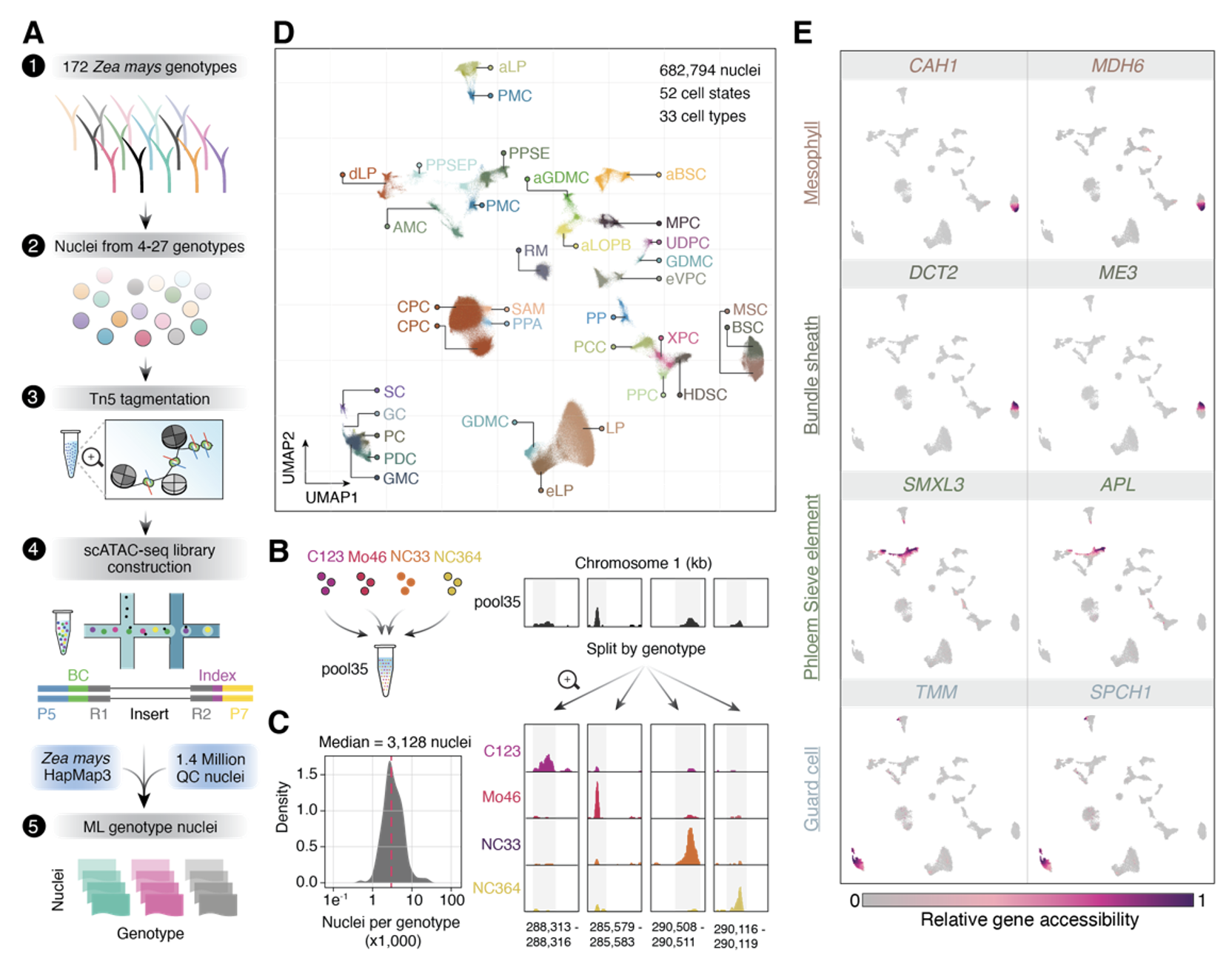
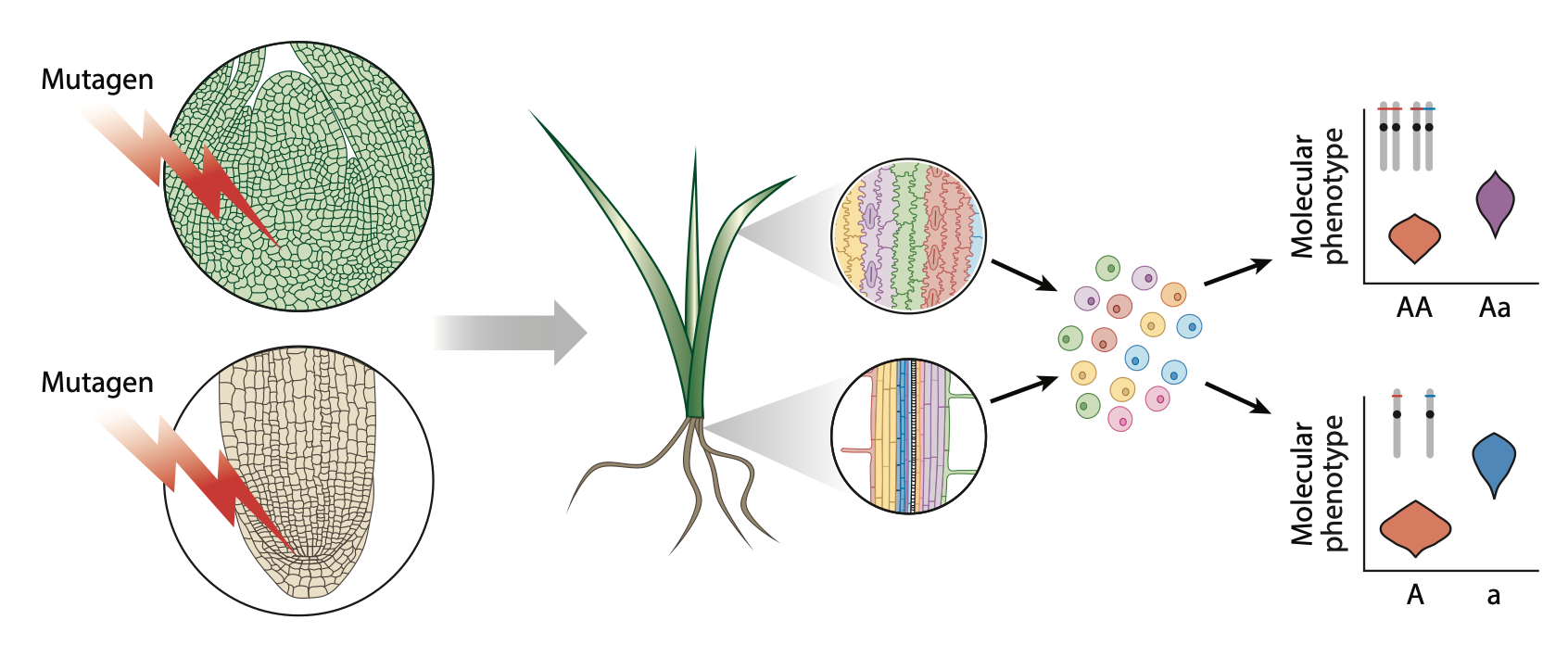
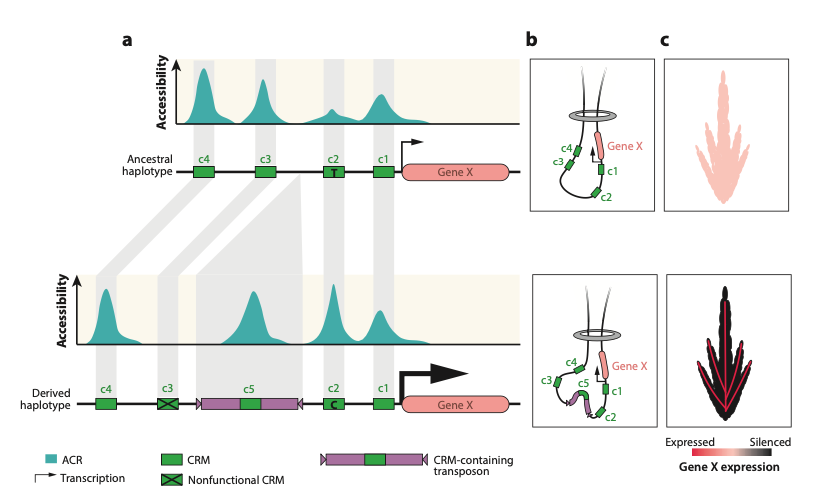
33. The genetic architecture of cell-type-specific cis-regulation in maize
Alexandre P. Marand#, Luguang Jiang, Fabio A. Gomez-Cano, Mark A. A. Minow, Xuan Zhang, John P. Mendieta, Ziliang Luo, Sohyun Bang, Haidong Yan, Cullan Meyer, Luca Schlegel, Frank Johannes, Robert J. Schmitz# (2025)
Science
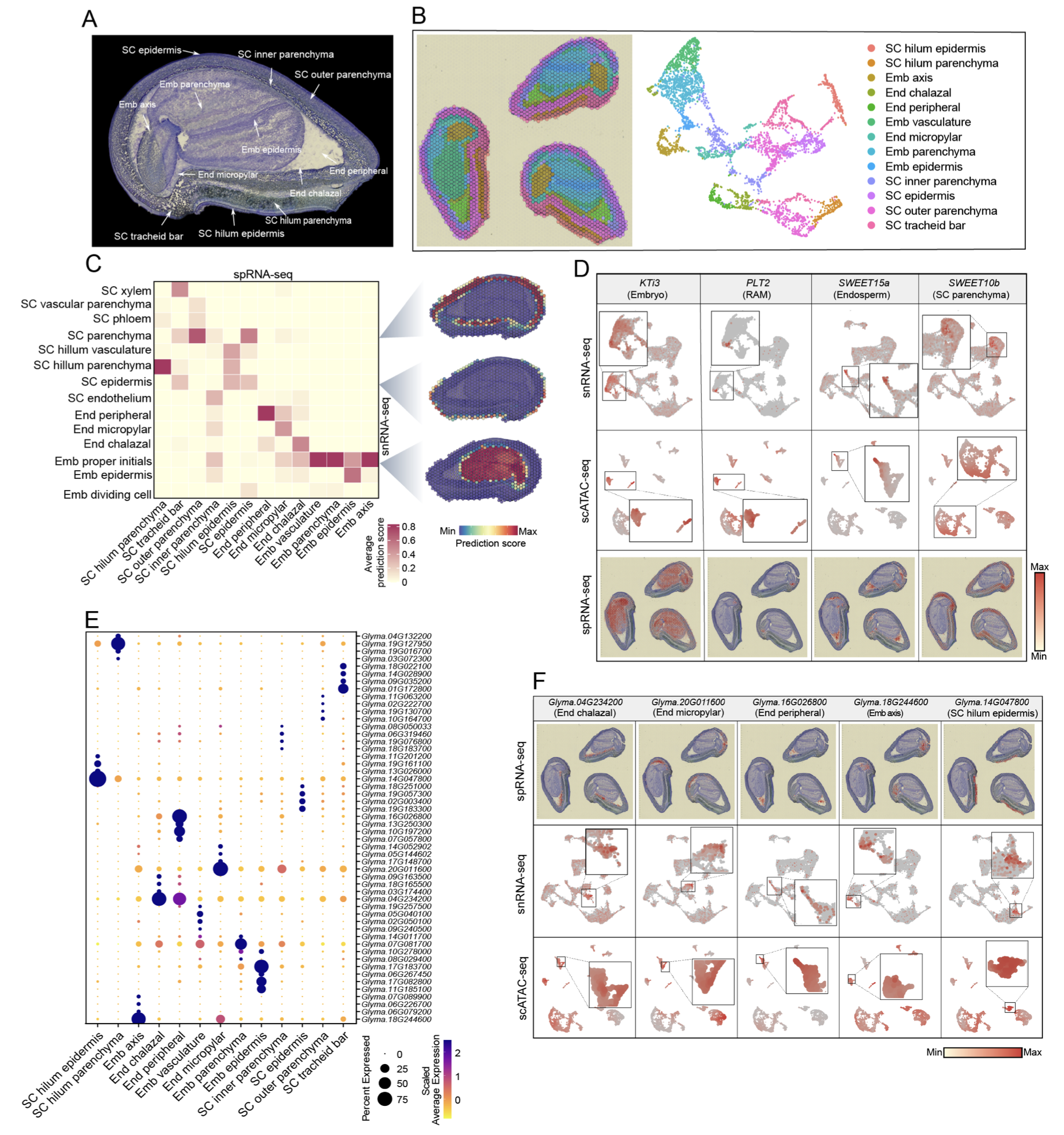
32. A spatially resolved multiomic single-cell atlas of soybean development
Xuan Zhang*, Ziliang Luo*, Alexandre P. Marand*, Haidong Yan, Hosung Jang, Sohyun Bang, John P. Mendieta, Mark A.A. Minow, Robert J. Schmitz (2025)
Cell
Check out the interactive Soybean Single Cell Spatial Omics Atlas Database
2024
31.Investigating the cis-Regulatory Basis of C3 and C4 Photosynthesis in Grasses at Single-Cell Resolution
John P. Mendieta, Xiaoyu Tu, Daiquan Jiang, Haidong Yan, Xuan Zhang, Alexandre P. Marand, Silin Zhong, Robert J. Schmitz (2024)
Proceedings of the National Academy of Sciences
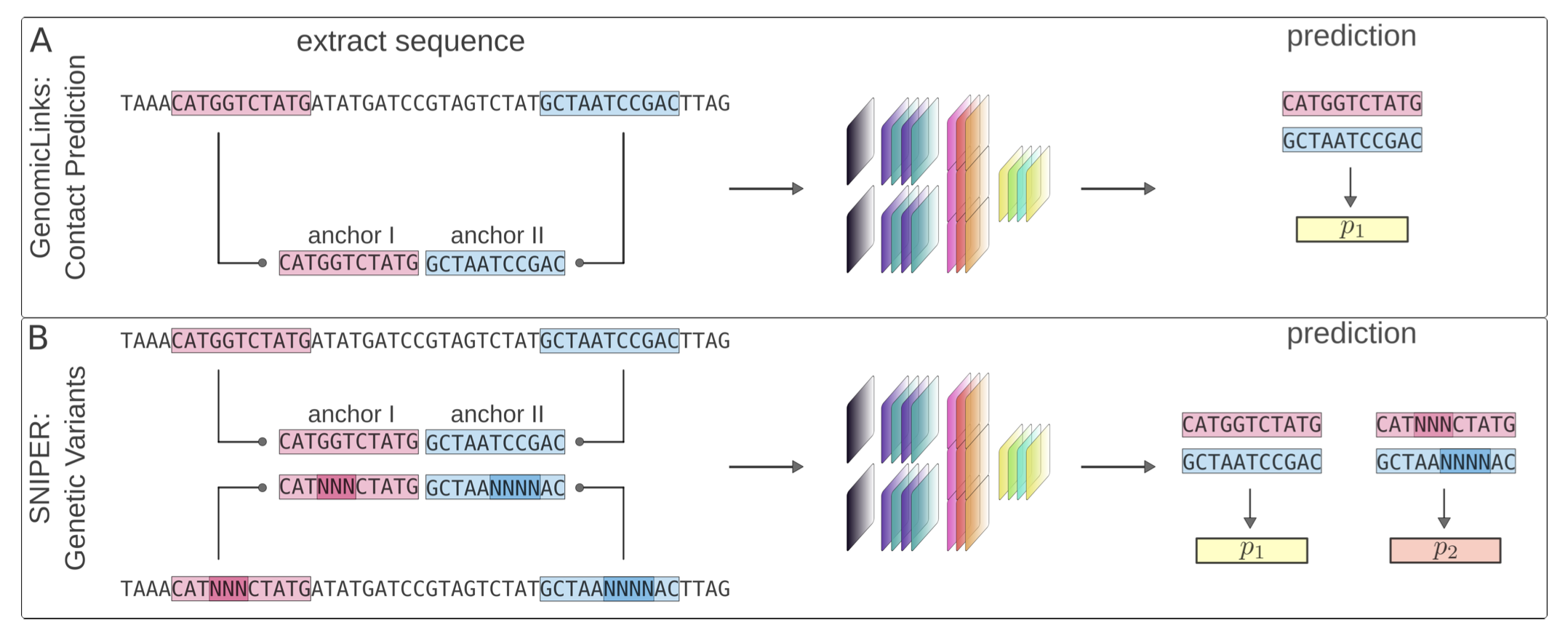
30. GenomicLinks: Deep learning predictions of 3D chromatin loops in the maize genome
Luca Schlegel, Rohan Bhardwaj, Yadollah Shahryary, Defne Demirtürk, Alexandre P. Marand, Robert J. Schmitz, Frank Johannes (2024)
Nucleic Acids Research Genomics and Bioinformatics
29. scifiATAC-seq: Massive-scale single-cell chromatin accessibility sequencing using combinatorial fluidic indexing
Xuan Zhang, Alexandre P. Marand, Haidong Yan, Robert J. Schmitz (2024)
Genome Biology
28. Computational Analysis of Maize Enhancer Regulatory Elements Using ATAC-STARR-seq
Alexandre P. Marand# (2024)
bio-protocol
2023
27. Leveraging Single-Cell Populations to Uncover the Genetic Basis of Complex Traits
Mark A.A. Minow, Alexandre P. Marand, Robert J. Schmitz (2023)
Annual Review of Genetics
26. cis-Regulatory Elements in Plant Development, Adaptation, and Evolution
Alexandre P. Marand, Andrea L. Eveland, Kerstin Kaufmann, Nathan Springer (2023)
Annual Review of Plant Biology
2022
25. Machine learning reveals conserved chromatin patterns determining meiotic recombination sites in plants
Minghui Wang, Shay Shilo, Adele Zhou, Mateusz Zelkowski, Mischa A. Olson, Ido Azuri, Nurit Shoshani-Hechel, Cathy Melamed-Bessudo, Alexandre P. Marand, Jiming Jiang, James C. Schnable, Charles J. Underwood, Ian R. Henderson, Qi Sun, Jaroslaw Pillardy, Penny M.A. Kianian, Shahryar F. Kianian, Changbin Chen, Avraham A. Levy, Wojciech P. Pawlowski. (2022)
bioRxiv
24. Modeling chromatin state from sequence across angiosperms using recurrent convolutional neural networks
Travis Wrightsman, Alexandre P. Marand, Peter A. Crisp, Nathan M. Springer, Edward S. Buckler. (2022)
The Plant Genome
23. A combinatorial indexing strategy for low cost epigenomic profiling of plant single cells
Xiaoyu Tu*, Alexandre P. Marand*, Robert J. Schmitz, Silin Zhong. (2022)
Plant Communications
22. Single-cell analysis of cis-regulatory elements
Alexandre P. Marand, Robert J. Schmitz. (2022)
Current Opinion in Plant Biology
2021
21. Quality control and evaluation of plant epigenomics data
Robert J. Schmitz, Alexandre P. Marand, Xuan Zhang, Rebecca A. Mosher, Franziska Turck, Xuemei Chen, Michael J. Axtell, Xuehua Zhong, Siobhan M. Brady, Molly Megraw, Blake C. Myers. (2021)
The Plant Cell
20. Vision, challenges, and opportunities for a Plant Cell Atlas
Plant Cell Atlas Consortium, Suryatapa Ghosh Jha, Alexander T. Borowsky, Benjamin J. Cole, Noah Fahlgren, Andrew Farmer, Shao-shan Carol Huang, Purva Karia, Marc Libault, Nicholas J. Provart, Selena L. Rice, Maite Saura-Sanchez, Pinky Agarwal, Amir H. Ahkami, Christopher R. Anderton, Steven P. Briggs, Jennifer A.N. Brophy, Peter Denolf, Luigi F. Di Costanzo, Moises Exposito-Alonso, Stefania Giacomello, Fabio Gomez-Cano, Kerstin Kaufmann, Dae Kwan Ko, Sagar Kumar, Andrey V. Malkovskiy, Naomi Nakayama, Toshihiro Obata, Marisa S. Otegui, Gergo Palfalvi, Elsa H. Quezada-Rodríguez, Rajveer Singh, R. Glen Uhrig, Jamie Waese, Klaas Van Wijk, R. Clay Wright, David W. Ehrhardt, Kenneth D. Birnbaum, Seung Y. Rhee. (2021)
eLife
19. Profiling single-cell chromatin accessibility in plants
Alexandre P. Marand, Xuan Zhang, Julie Nelson, Pedro Augusto Braga dos Reis, Robert J. Schmitz. (2021)
STAR Protocols
18. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes
Matthew B. Hufford, Arun S. Seetharam, Margaret R. Woodhouse, Kapeel M. Chougule, Shujun Ou, Jianing Liu, William A. Ricci, Tingting Guo, Andrew Olson, Yinjie Qiu, Rafael Della Coletta, Silas Tittes, Asher I. Hudson, Alexandre P. Marand, Sharon Wei, Zhenyuan Lu, Bo Wang, Marcela K. Tello-Ruiz, Rebecca D. Piri, Na Wang, Dong won Kim, Yibing Zeng, Christine H. O’Connor, Xianran Li, Amanda M. Gilbert, Erin Baggs, Ksenia V. Krasileva, John L. Portwood II, Ethalinda K.S. Cannon, Carson M. Andorf, Nancy Manchanda, Samantha J. Snodgrass, David E. Hufnagel, Qiuhan Jiang, Sarah Pedersen, Michael L. Syring, David A. Kudrna, Victor Llaca, Kevin Fengler, Robert J. Schmitz, Jeffrey Ross-Ibarra, Jianming Yu, Jonathan I. Gent, Candice N. Hirsch, Doreen Ware, R. Kelly Dawe. (2021)
Science
17. Leveraging histone modifications to improve genome annotations
John P. Mendieta, Alexandre P. Marand, William A. Ricci, Xuan Zhang, Robert J. Schmitz. (2021)
G3 Genes|Genomes|Genetics
16. Stability of DNA methylation and chromatin accessibility in structurally diverse maize genomes
Jaclyn M. Noshay, Zhikai Liang, Peng Zhou, Peter A. Crisp, Alexandre P. Marand, Candice N. Hirsch, Robert J. Schmitz, Nathan M. Springer. (2021)
G3 Genes|Genomes|Genetics
15. A cis-regulatory atlas in maize at single-cell resolution
Alexandre P. Marand#, Zongliang Chen, Andrea Gallavotti, Robert J. Schmitz#. (2021)
Cell
Coverage:
PNAS news
Plantae
UGA Research
Cell preview
14. Chromatin accessibility profiling methods
Liesbeth Minnoye, Georgia K. Marinov, Thomas Krausgruber, Lixia Pan, Alexandre P. Marand, Stefano Secchia, William J. Greenleaf, Eillen E. M. Furlong, Keigi Zhao, Robert J. Schmitz, Christoph Bock, Stein Aerts. (2021)
Nature Reviews Methods Primers
2020
13. Assessing the regulatory potential of transposable elements using chromatin accessibility profiles of maize transposons
Jaclyn M. Noshay, Alexandre P. Marand, Sarah N. Anderson, Peng Zhou, Maria Katherine Mejia Guerra, Zefu Lu, Christine O’Connor, Peter A. Crisp, Candice N. Hirsch, Robert J. Schmitz, Nathan M. Springer. (2020)
Genetics
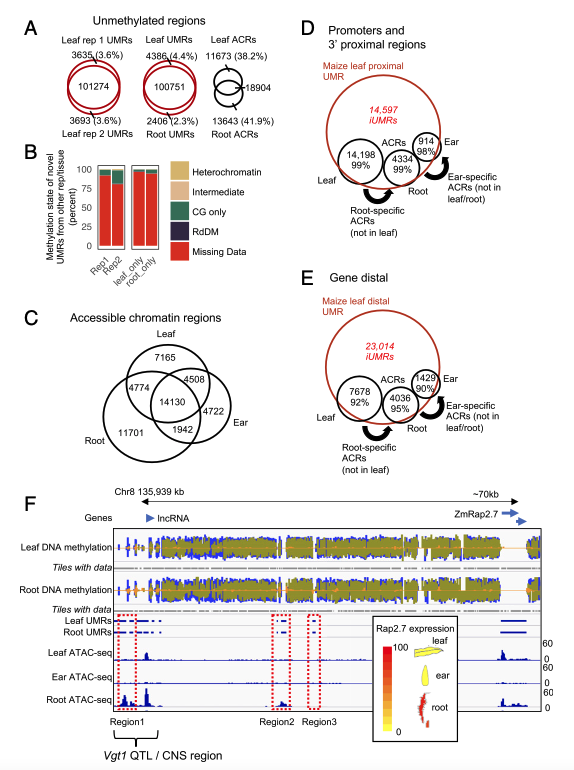
12. Stable unmethylated DNA demarcates expressed genes and their cis-regulatory space in plant genomes
Peter A. Crisp, Alexandre P. Marand, Jaclyn M. Noshay, Peng Zhou, Zefu Lu, Robert J. Schmitz, Nathan M. Springer. (2020)
Proceedings of the National Academy of Sciences
11. Identification of a novel base J binding protein complex involved in RNA polymerase II transcription termination in trypanosomes
Rudo Kieft, Yang Zhang, Alexandre P. Marand, Jose Dagoberto Moran, Robert Bridger, Lance Wells, Robert J. Schmitz, Robert Sabatini. (2020)
PLOS Genetics
2019
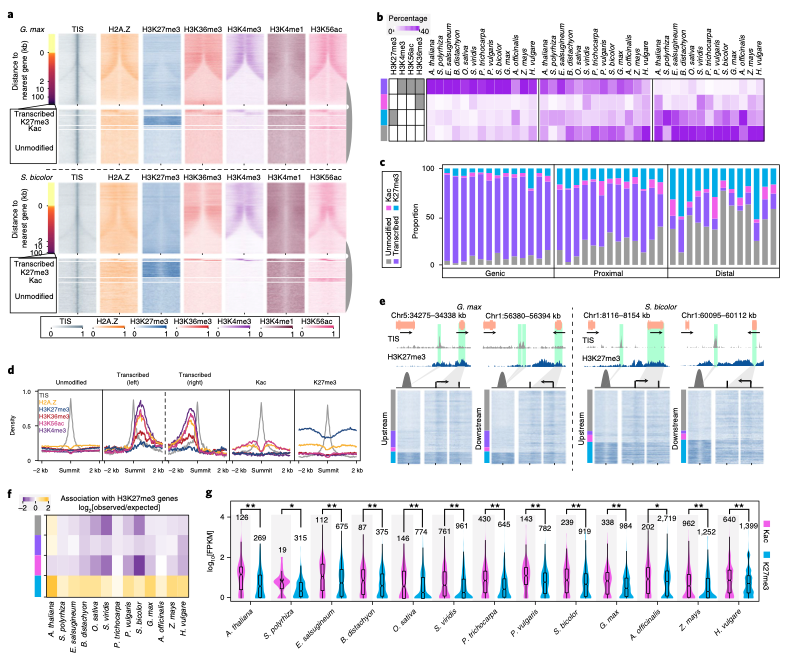
10. The prevalence, evolution and chromatin signatures of plant regulatory elements
Zefu Lu, Alexandre P. Marand, William A. Ricci, Christina L. Ethridge, Xiaoyu Zhang, Robert J. Schmitz. (2019)
Nature Plants
9. Widespread long-range cis-regulatory elements in the maize genome
William A. Ricci*, Zefu Lu*, Lexiang Ji*, Alexandre P. Marand, Christina L. Ethridge, Nathalie G. Murphy, Jaclyn M. Noshay, Mary Galli, María Katherine Mejía-Guerra, Maria Colomé-Tatché, Frank Johannes, M. Jordan Rowley, Victor G. Corces, Jixian Zhai, Michael J. Scanlon, Edward S. Buckler, Andrea Gallavotti, Nathan M. Springer, Robert J. Schmitz, Xiaoyu Zhang. (2019)
Nature Plants
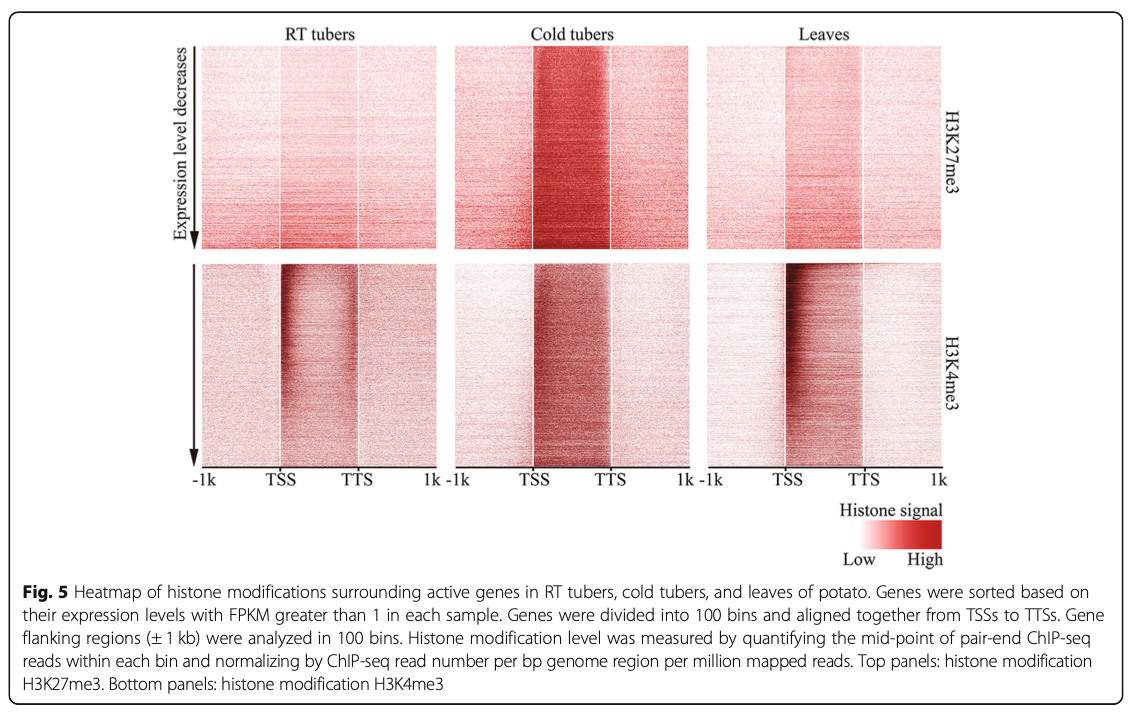
8. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato
Zixian Zeng, Wenli Zhang, Alexandre P. Marand, Bo Zhu, C. Robin Buell, Jiming Jiang. (2019)
Genome Biology
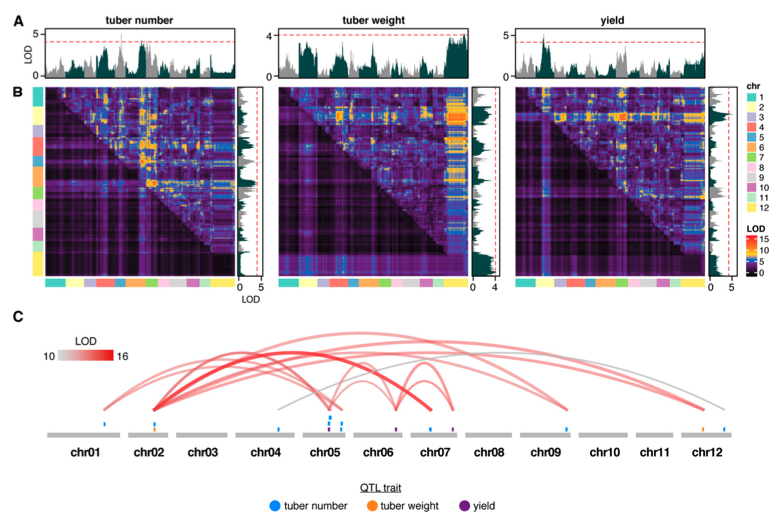
7. Residual Heterozygosity and Epistatic Interactions Underlie the Complex Genetic Architecture of Yield in Diploid Potato
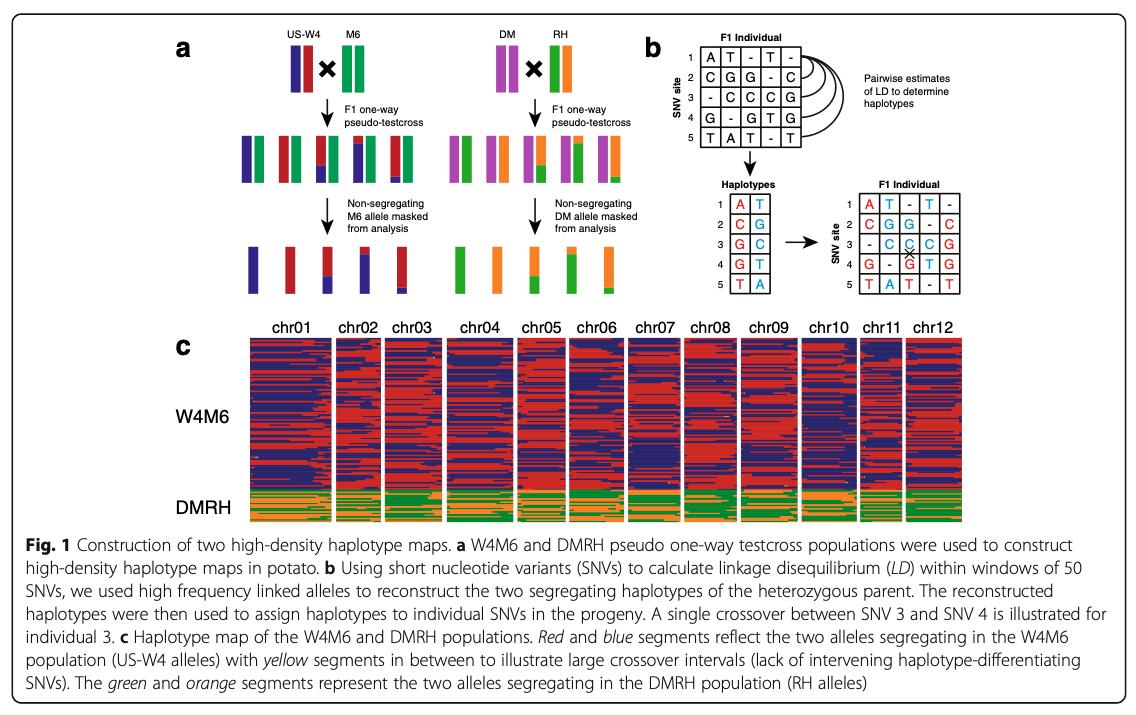
Alexandre P. Marand, Shelley H. Jansky, Joseph L. Gage, Andy J. Hamernik, Natalia de Leon, Jiming Jiang. (2019)
Genetics
6. Historical Meiotic Crossover Hotspots Fueled Patterns of Evolutionary Divergence in Rice
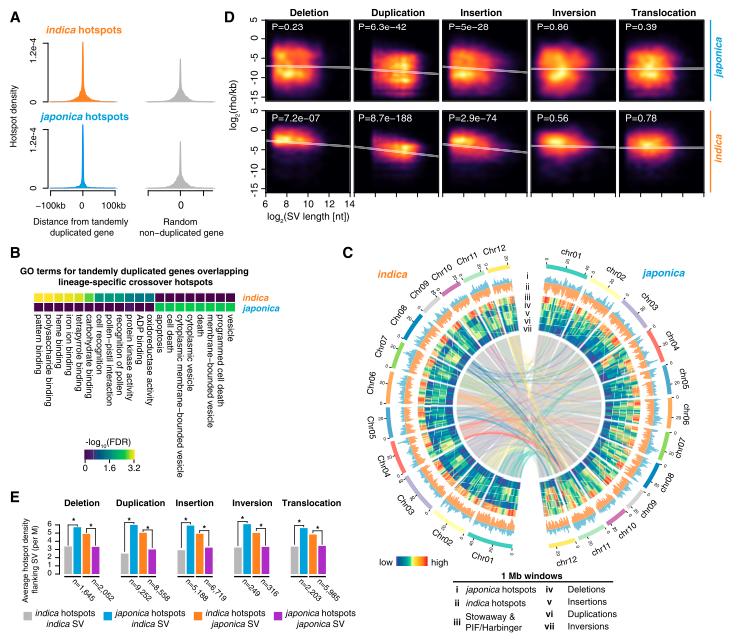
Alexandre P. Marand, Hainan Zhao, Wenli Zhang, Zixian Zeng, Chao Fang, Jiming Jiang. (2019)
The Plant Cell
2018
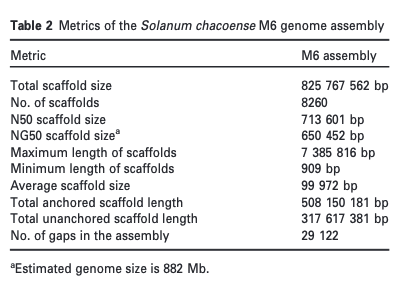
5. Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity
Courtney P. Leisner, John P. Hamilton, Emily Crisovan, Norma C. Manrique-Carpintero, Alexandre P. Marand, Linsey Newton, Gina M. Pham, Jiming Jiang, David S. Douches, Shelley H. Jansky, C. Robin Buell. (2018)
The Plant Journal
4. Proliferation of regulatory DNA elements derived from transposable elements in the maize genome
Hainan Zhao, Wenli Zhang, Lifen Chen, Lei Wang, Alexandre P. Marand, Yufeng Wu, Jiming Jiang. (2018)
Plant Physiology
2017
3. Meiotic crossovers are associated with open chromatin and enriched with Stowaway transposons in potato
Alexandre P. Marand, Shelley H. Jansky, Hainan Zhao, Courtney P. Leisner, Xiaobiao Zhu, Zixian Zeng, Emily Crisovan, Linsey Newton, Andy J. Hamernik, Richard E. Veilleux, C. Robin Buell, Jiming Jiang. (2017)
Genome Biology
2. Towards genome-wide prediction and characterization of enhancers in plants
Alexandre P. Marand, Tao Zhang, Bo Zhu, Jiming Jiang. (2017)
BBA Gene Regulatory Mechanisms
2016
1. PlantDHS: a database for DNase I hypersensitive sites in plants
Tao Zhang, Alexandre P. Marand, Jiming Jiang. (2016)
Nucleic Acids Research