Motivation
The long term goal of the lab is to understand how the genome of a single cell reproducibly encodes the development and morphology of an entire multicellular organism. Cellular diversity within and between organisms is largely attributed to differences in transcriptional regulation, with cis-regulatory elements - sequences that control when and where a gene is transcribed - playing a central role. To tackle this question, the lab is currently focused on three major objectives:
-
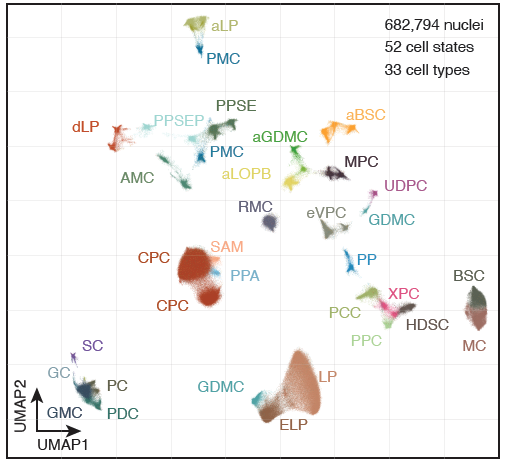
Identify the comprehensive catalog of cis-regulatory elements and their associated trans-factors underlying diverse cell identities and fate transitions in plant genomes. Leveraging recent advances in single-cell profiling methods, the lab primarily focuses these efforts in Zea mays and Arabidopsis thaliana and aims to expand to other model systems.
-
Development of new techniques (experimental and computational) for quantitatively assaying non-coding regulatory element function. Efforts to chart the locations and functions of silencers, chromatin organization elements, and other cis-regulatory types have remained relatively unexplored compared to enhancers and promoters, including in animal systems. Understanding how different combinations of regulatory elements result in a specific quantitative activity (and outcomes on transcription) is a major objective of the lab.
-
Determining how (epi)genetic and (epi)genomic variation affects cis-regulatory activity and transcription regulation at the molecular level. Coupling standing genetic variation and mutagenesis approaches with high-throughput sequencing and reporter assays at the single-cell level facilitates characterization of novel molecular mechanisms underlying plasticity in cellular function, identity, and development.
Single-cell genomics for decoding development
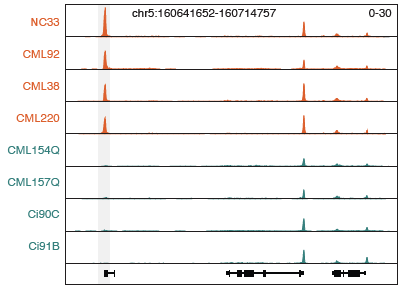
A major goal of developmental biology is elucidating how morphological and developmental variation arises within a species. Accumulating evidence from Genome-wide Association Studies implicates genetic variation within cis-regulatory elements as a significant source affecting phenotypic traits. Current paradigms suggest that regulatory variation may affect chromatin structure and transcription factor binding that result in altered patterns of transcription that encourage phenotypic evolution. Results from the lab indicate significant enrichment of phenotype-associated genetic variants within cell-type-specific cis-regulatory elements, suggesting that trait-associated variation are affecting transcriptional regulation in specific cellular contexts.
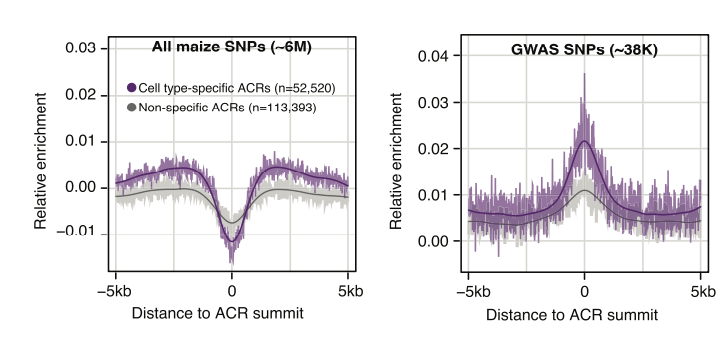
To better chart the genetic determinants of regulatory function, we are using single-cell genomics to profile chromatin accessibility in diverse genetic backgrounds in maize. This project aims to identify genetic variants affecting levels of chromatin accessibility in diverse cellular contexts, their effects on transcript abundance, and the consequences on cellular and organism-scale phenotypes. A comprehensive map of molecular effects will enable future genome engineering efforts to predictably fine-tune important traits.